Are Mrna Sequences Read 3' to 5'
Let the states brand an in-depth study of the RNA processing. After reading this article you volition learn nigh: 1. Processing of Eukaryotic mRNA ii. RNA Splicing and Mechanisms of Splicing and 3. Processing of tRNA.
Almost all types of RNA molecules undergo post synthesis transformation which is called RNA processing. Prokaryotic mRNA is generally non processed. In prokaryotes v′-end of prokaryotic mRNA starts translation while the 3′-terminate is still under synthesis. Eukaryotic mRNA undergoes maximum processing. Both prokaryotic and eukaryotic tRNAs and rRNAs undergo processing. Except prokaryotic mRNA, all other kinds of RNA are candy immediately after synthesis.
Processing of Eukaryotic mRNA:
Newly synthesized mRNA is called primary transcription or precursor mRNA. It is quite different from the mRNA that takes part in protein synthesis. Large-scale changes take place in precursor mRNA. These changes are chosen processing of mRNA. Both 5′-end three′-end of mRNA are modified. Non-coding regions are removed by splicing. The changes lead to the formation of mature mRNA which takes part in protein synthesis.
v′-cease Capping:
At five′-end of mRNA a cap of 7-methylguanosine (grand7G or 7MeG) is added immediately afterwards transcription or during transcription. This is nearly common and is called a cap. This cap is added in contrary orientation of five′ → 5′ instead of normal five′ → 3′ orientation giving a five-5′ triphosphate span. A methyl group is added to the 7thursday position of terminal guanine. This reaction is catalysed by an enzyme guanylyl transferase. The cap provides stability to mRNA as the exonuclease is unable to act on information technology.
three′-Cleavage and Polyadenylation:
At the 3′-end of mRNA a tail consisting of sequence of adenine (adenylic acid) is added immediately after transcription. This tail is chosen poly (A) tail. It consists of 80-200 adenine nucleotides. This addition is catalysed by an enzyme poly (A) polymerase or PAP. The tail provides stability to mRNA.
RNA Splicing and Mechanisms of Splicing:
Till recently it was believed that coding sequences of Dna and amino acids of polypeptide chain are collinear. Recently it has been discovered that coding sequences of most of the eukaryotic genes is split into stretches of codons interrupted past stretches of not-coding sequences.
The coding sequences of DNA are called exons. In between exons, there are intervening non-coding sequences called introns. This type of genes are chosen divide genes or interrupted genes. The terms exons and introns were given past Gilbert. Introns are usually much larger than the exons. Moreover, the introns plant a large portion of the genome. During processing of mRNA, the introns are excised out and removed past a process called RNA splicing. Translation takes identify after the splicing is completed.
Processing of tRNA:
tRNA undergoes extensive processing. The mature tRNAs consists of lxxx-90 nucleotides. But the precursor tRNA is much longer. For example tRNATyr which is a tyrosine conveying tRNA contains 350 nucleotides. Processing discards useless sequences. This is done by enzymes RNase D, RNase Due east, RNase F and RNase P. Nucleotides are removed from both 5′ and 3′ ends.
Endonucleases besides remove many sequences. Cleaving is done after the primary transcript has folded and formed characteristic stems and loop structure by extensive complementary base pairing. RNase P is a ribozyme.
The 5′-CCA-3′ sequence at 3′-stop of mature tRNA is added by the enzyme tRNA nucleotidyl transferse. This generates the mature 3′-end the tRNA.
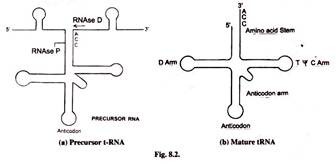
Several unusual bases are formed past the modification of normal existing bases A, G, C and U by the enzymatic action. These modified bases are pseudouridine (Ψ), 2- isopentenyladenosine (ii ip A), ii-O-methylguanosine (2m G), 4-thiouridine (4 t μ), Ribothymidine, dihyrouridine and inosine.
rRNA Processing:
In rRNA processing in both prokaryotes and eukaryotes, the primary transcript undergoes some changes. Some nucleotides are removed by exonucleases and endonucleases. New nucleotides are added both at v′-end and 3′-end. Certain nucleotides are modified.
rRNA Processing in Prokaryotes:
In East. coli there are several dissimilar operons in rRNA. Each operon contains one copy of 5S, 16S and 23S rRNA sequences. In addition, several coding sequences of tRNA molecules are also present in rRNA operons. These main transcriptions are candy to give both rRNA and tRNA molecules. All these molecules are broken from a continuous transcript of more than than 5000 nucleotides.
rRNA Processing in Eukaryotes:
The precursor rRNA in eukaryotes comprise one re-create of 18S coding region, one re-create of 5.8S and one copy of 28S coding region. This rRNA precursor is transcribed in the nucleolus past RNA polymerase I. The eukaryotic 5S rRNA is transcribed by RNA polymerase III from an unlinked gene.
In both prokaryotes and eukaryotes rRNA molecules class secondary structures of numerous double stranded stems past complementary base pairing and single stranded loops.
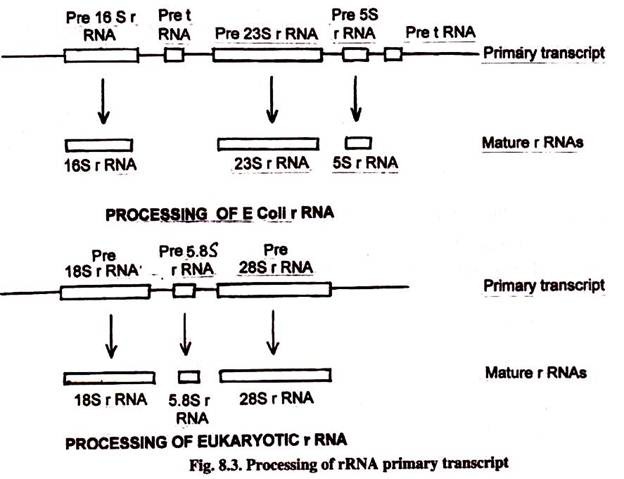
Small nucleolar RNA (snoRNA) is required for the processing of rRNA molecules in eukaryotes. The snoRNAs are nowadays in the nucleolus where rRNA is processed. Ribosomes are also assembled in nucleolus.
The RNA molecules in cells are present in the class of complexes formed with proteins. Specific proteins bind to specific RNAs. The RNA-protein complexes are called ribonuclear proteins or RNPs. Ribosomes are the largest RNPs formed by rRNA molecules forming complexes with specific ribosomal proteins. In E. coli ribosomes business relationship for 25% of the dry weight of the jail cell. Information technology consists of 10% of total proteins and 80% of full RNA.
In prokaryotes 70S ribosome has 30S and 50S subunits. The 30S subunit has one re-create of 16S rRNA and 21 different proteins. The 50S subunit has 23S rRNA, 5S rRNA and 31 different proteins.
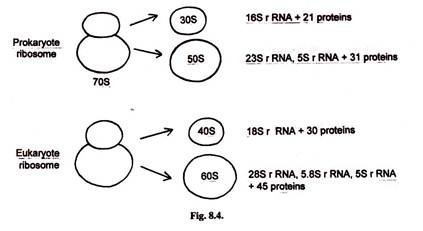
In eukaryotes 80S ribosome has 40S and 60S subunits. The 40S subunit has 18S rRNA and thirty different proteins. The 60S subunit has 28S rRNA, v.8S rRNA, 5.8S rRNA and 45 different proteins.
Summary :
Except prokaryotic mRNA all other kinds of RNAs are processed immediately later on synthesis. Eukaryotic mRNA undergoes maximum processing. Both prokaryotic arid eukaryotic tRNAs and rRNAs undergo extensive processing. Processing of eukaryotic mRNA: Newly synthesized mRNA is called pre-mRNA. It is quite different from the mRNA that takes role in protein synthesis. At 5′ end of mRNA, a cap of 7-methylguanosine (yardsevenG or 7 Me M) is added. At iii′-end a tail consisting of lxxx-200 adenine nucleotides called Poly(A) tail is added.
Not-coding regions or introns are removed by splicing and coding regions or exons are joined together.
Processing of tRNA:
Pre-tRNA consists of upto 350 nucleotides. Processing discards most of useless nucleotides. Mature tRNA consists of about 70 nucleotides. At the 3′ finish CCA sequence is added. Several unusual bases are constitute by the modification of normal bases A, G, C and U.
rRNA Processing:
In both prokaryotes and eukaryoties rRNA molecules form secondary structures of numerous double stranded stems by complementary base pairing and single stranded loops. Many proteins bind to these rRNA molecules to form ribonucleoproteins (RNP) in the ribosomes. In that location are several pocket-size nuclear RNA molecules chosen small nuclear RNAs (snRNA).
Source: https://www.biologydiscussion.com/rna/rna-processing-eukaryotic-mrna-and-trna-processing-with-diagram/16381
0 Response to "Are Mrna Sequences Read 3' to 5'"
Post a Comment